PSA Database Handbook
Last Updated: 06/05/2020 by Aurelie
-
Nomenclature and Definition
Tables:

Constraints:

Missing Data:
- NA if data is missing because data collection is ongoing.
- -999 (for numbers and text) and 1900-01-01 (for dates) if data collection is over and data is missing.
Naming Convention:
- All table names and columns are in lowercase.
- Words in table and column names were delimited using underscores.
- Date-time format is alway timestamp without time zone and time zone is always GMT.
- All coordinates are expressed in the WGS84 latitude, longitude format, with latitude and longitude data being stored in two different columns.
-
Site Information and Producer IDs
Description:
-
Data in the
producer_idsandsite_informationtables are collected using the tech dashboard. New rows will be added to these tables during onboarding. Existing rows will be updated by the data creator directly from the tech dashboard without requiring approval of a data shepherd. There are no validation columns associated with these two tables. -
When onboarding a new site, the user will have the choice between creating a new producer (or partner) or updating the contact information of an existing producer (or partner). New codes will be automatically associated to a site during onboarding.
-
Unique producer IDs are attributed using the following format:
yyyysssssxxxwithyyyy: year of entry in network, specified using 4 digits;sssss: affiliated abbreviation (up to 5 characters); andxxx: up to 3-digit numbers. -
For PSA on-farm sites: Site codes are defined using 3 letters.
-
For partner sites: Site codes are defined using 2 letters plus 1 digit ranging from 1 to 9 (zeros were excluded to avoid confusions with the letter O).
-
Data strings stored in the code, affiliation, and county columns are all uppercase.
-
The
collaboration_statustable and collaboration_status column in theaffiliationstable states if the producer is affiliated with a university or an industry partner.
Relational Diagram:
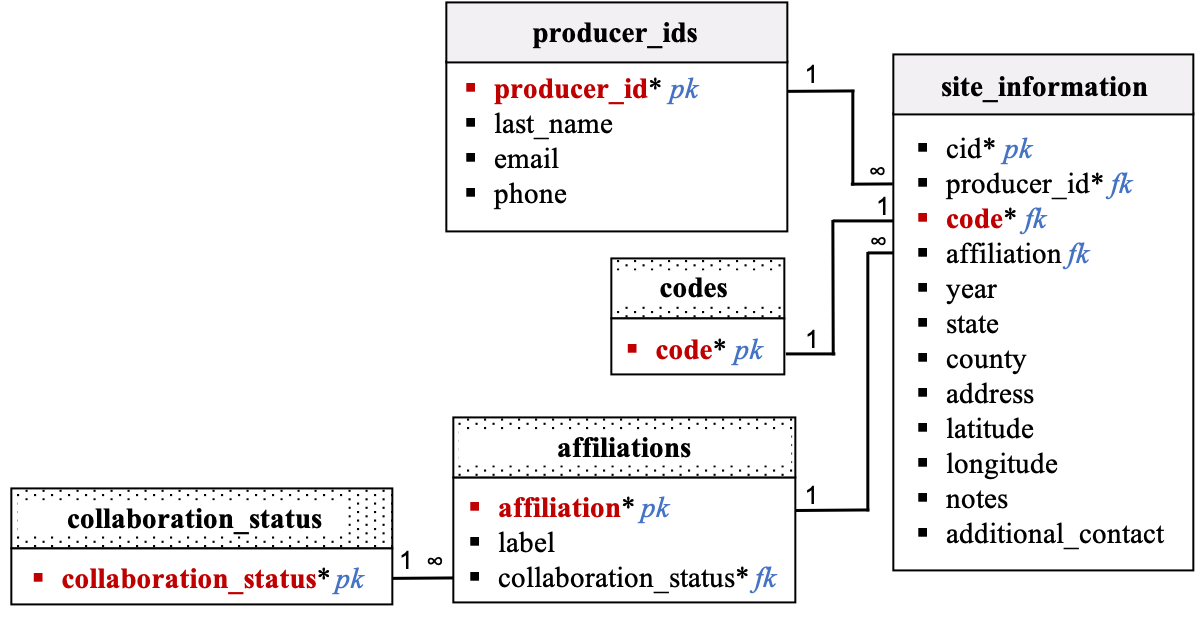
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
collaboration_status:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| collaboration_status | charvar(15) | NA | University or industry partner. |
affiliations:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| affiliation | charvar(25) | NA | Lists all states and partners abbreviations. |
| label | charvar(50) | NA | Labels corresponding to states and partners abbreviations. |
| collaboration_status | charvar(15) | NA | University or industry partner. |
producer_ids:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| producer_id | char(9) | NA | Unique producer ID. |
| last_name | charvar(20) | NA | Grower’s last name. |
| charvar(50) | NA | Grower’s up-to-date email. | |
| phone | char(10) | NA | Grower’s up-to-date phone number. |
site_information:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row ID (ignore). |
| code | char(3) | NA | Site code. |
| producer_id | char(9) | NA | Producer ID associated to site code. |
| affilitation | char(25) | NA | Affiliated state or partner. |
| year | char(4) | NA | Year of the experiment. |
| state | char(2) | NA | State. |
| county | charvar(30) | NA | County. |
| address | text | NA | Address of experimental site. |
| latitude | real | decimal degree | Latitude of experimental site [WGS84]. |
| longitude | real | decimal degree | Longitude of experimental site [WGS84]. |
| notes | text | NA | Optional notes. |
| additional_contact | text | NA | Optional additional contact. |
-
Protocol Enrollment
Description:
-
The
protocol_statustable lists all the existing PSA on-farm protocols and defines which protocols are currently active within the on-farm network. This table will need to be updated when a new protocol is being created and launched within the network, and when an existing protocol is being retired. -
The
protocol_enrollmenttable lists which protocols were/will be conducted at every site. New rows will be created when a new site is onboarded and data in this table will be completed when the field team selects protocols (by site) in the tech dashboard. The field team should only be able to see and select active protocols in the tech dashboard. New columns will need to be added when new protocols are launched. -
There are no validation columns associated with these two tables and the field team will be able to update information provided in
protocol_enrollmentdirectly from the tech dashboard without requiring approval of a data shepherd. Theprotocol_statustable will not be accessible to the field team via the tech dashboard. -
The data flow team needs to think about how new protocols will be added to these two tables (new rows in
protocol_statusand new columns inprotocol_enrollment). -
In the
protocol_enrollmenttable, default values in the following columns: soil_nitrogen and bulk density are set to zero because these protocols have been retired. The data flow team will need to remember setting default values to zeros when retiring a protocol. Default values in the columns: farm_history were set to one because that protocol is mandatory and information must be collected for all PSA on-farm sites. -
The data flow team will have to think about the steps that need to be taken when a field team member select a protocol that was not initially selected during onboarding. Indeed if a protocol is not selected at onboarding, tables will still be pre-filled, but they will be pre-filled with missing values and these will need to be updated to null values (and zeros in the validation columns).
Relational Diagram:
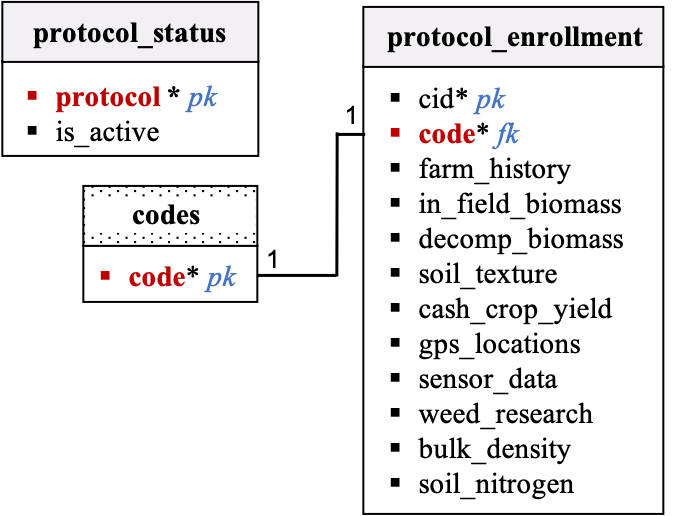
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
protocol_status:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| protocol | charvar(30) | NA | Lists all existing PSA on-farm protocols. |
| is_active | integer | NA | Equals 1 if protocol is active, and 0 otherwise. |
protocol_enrollment:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| farm_history | integer | NA | Equals 1 if farm history data are collected at corresponding site, and 0 otherwise. |
| in_field_biomass | integer | NA | Equals 1 if in field biomass data are collected at corresponding site, and 0 otherwise. |
| decomp_biomass | integer | NA | Equals 1 if decomp biomass data are collected at corresponding site, and 0 otherwise. |
| soil_texture | integer | NA | Equals 1 if soil texture data are collected at corresponding site, and 0 otherwise. |
| cash_crop_yield | integer | NA | Equals 1 if cash crop yield data are collected at corresponding site, and 0 otherwise. |
| gps_locations | integer | NA | Equals 1 if plot corners and subplot GPS locations are recorded at corresponding site, and 0 otherwise. |
| sensor_data | integer | NA | Equals 1 if TDR sensor data are collected at corresponding site, and 0 otherwise. |
| weed_research | integer | NA | Equals 1 if weed pictures are collected at corresponding site, and 0 otherwise. |
| bulk_density | integer | NA | Equals 1 if surface bulk density measurements were collected at corresponding site, and 0 otherwise. |
| soil_nitrogen | integer | NA | Equals 1 if soil nitrogen core samples were collected at corresponding site, and 0 otherwise. |
-
Farm History
Description:
-
Data in the
farm_historytable will be collected using Kobo. There are validation columns in this table. The field team will be able to suggest edits in the tech dashboard and the designated data shepherd will update data as needed. GIT issue tracker will be used in the background to keep track of suggestions and data updates. -
Data validation will be performed for each site code. By default, values in the validation_by_creator and validation_by_shepherd columns are set to 0. After validation by the data creator and data shepherd, values in the validation_by_creator and validation_by_shepherd columns are set to 1. If
farm_historydata for a given code are validated by both the data creator and the data shepherd while some values are still missing, then all missing values will be converted to -999 or 1900-01-01. If allfarm_historydata is missing for a given site and data is validated by the data creator and shepherd, then all values are being converted to -999 or 1900-01-01 and values in the validation_by_creator and validation_by_shepherd columns for that code are set to -999. The farm_history protocol is mandatory, and if values in the validation_by_creator and validation_by_shepherd columns forfarm_historyare set to -999, we consider that this particular site is not part of the study anymore. -
Rows in the
farm_historytable will be pre-filled with empty values (and zeros in the validation columns) at onboarding. One row should be created for each farm code. -
kill_time: equals -1 if cover crop was killed before cash crop planting. Equals 0 if cover crop was killed at cash crop planting. Equals 1 if cover crop was killed after cash crop planting.
Relational Diagram:
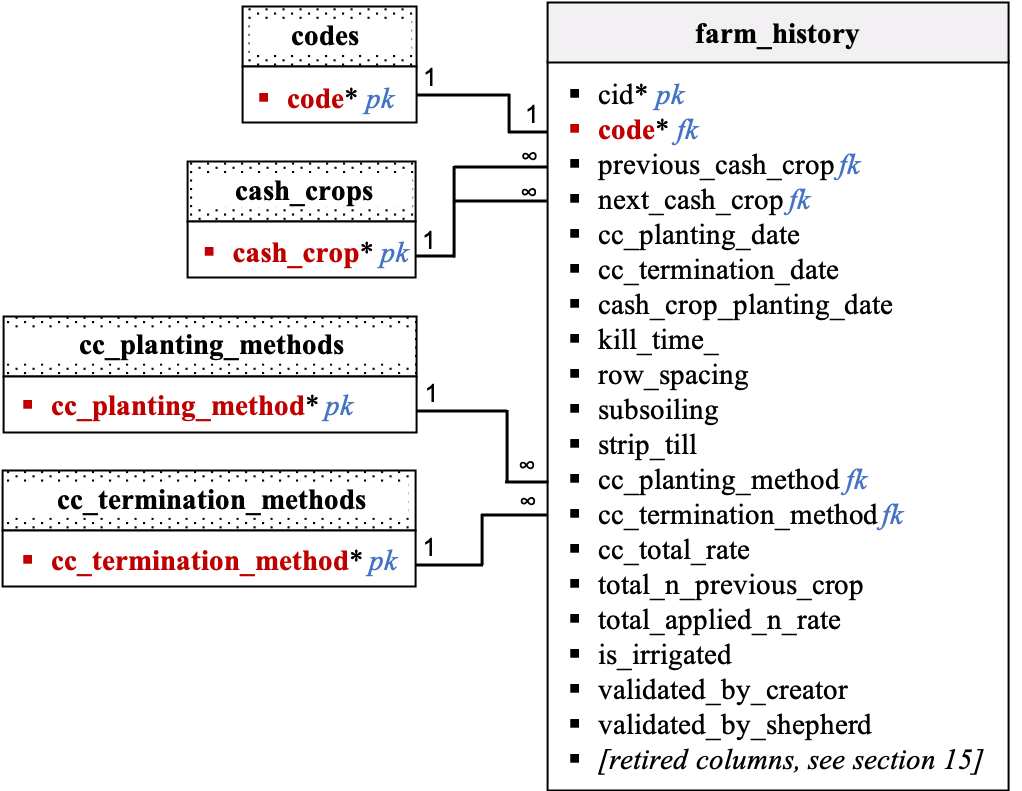
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
cash_crops:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cash_crop | charvar(20) | NA | Lists all cash crops. |
cc_planting_methods:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cc_planting_method | charvar(20) | NA | Lists all cover crop planting methods. |
cc_termination_methods:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cc_termination_methods | charvar(35) | NA | Lists all cover crop termination methods. |
farm_history:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| previous_cash_crop | charvar(20) | NA | Last cash crop grown prior to cover crop. |
| next_cash_crop | charvar(20) | NA | Cash crop grown after cover crop. |
| cc_planting_date | date | NA | Cover crop planting date. |
| cc_termination_date | date | NA | Cover crop termination date. |
| cash_crop_planting_date | date | NA | Cash crop planting date. |
| kill_time | integer | NA | Cover crop kill time relatively to cash crop planting. |
| row_spacing | real | in | Cash crop row spacing. |
| subsoiling | integer | NA | Equals 1 if site was subsoiled, 0 otherwise. |
| strip_till | integer | NA | Equals 1 if site was strip-tilled, 0 otherwise. |
| cc_planting_method | charvar(20) | NA | Cover crop planting method. |
| cc_termination_method | charvar(35) | NA | Cover crop termination method. |
| cc_total_rate | integer | lbs/ac | Cumulative cover crop seeding rate. |
| total_n_previous_crop | text | NA | Total nitrogen applied to previous cash crop. |
| total_applied_n_rate | text | NA | Total nitrogen applied to following cash crop. |
| is_irrigated | integer | NA | Equals 1 if site was irrigated, 0 otherwise. |
| validated_by_creator | integer | NA | Defines validation by data creator. |
| validated_by_shepherd | integer | NA | Defines validation by data shepherd. |
-
Cover Crop Mixture and Applied Chemicals
Description:
-
The
cc_mixture,cc_species,cc_families, andcc_mixture_validationtables characterize the cover crop planted at each site. Theapplied_chemicals,chemical_names,chemical_families, andchemical_validationtables list the chemicals applied to each site (when applicable). Data in these tables will be collected using Kobo. After data collection, the field team will be able to suggest edits in the tech dashboard and the designated data shepherd will update data as needed. GIT issue tracker will be used in the background to keep track of suggestions and data updates. -
Validation columns are associated with the
cc_mixtureandapplied_chemicalstables. Because growers are free to plant any number of cover crops in their field and apply any combination of chemicals, validation of thecc_mixtureandapplied_chemicalsdata is completed in separate tables:cc_mixture_validationandchemical_validation, respectively. Validation will be performed by site code. By default, values in the validation_by_creator and validation_by_shepherd columns are set to 0. If at least one row/observation is recorded in thecc_mixtureandapplied_chemicalstables for a given site and that information was validated by the data creator and shepherd, then values of the validation_by_creator and validation_by_shepherd columns for that particular site are set to 1. If the data collection is ongoing or the data was not validated by the data creator and shepherd, then values of the validation_by_creator and validation_by_shepherd columns for that particular site remain equal to 0. If data is just missing for that particular site, and the missing data is being validated by the data creator and shepherd, then values of the validation_by_creator and validation_by_shepherd columns for that site are set to -999. -
Rows in the
cc_mixture_validationandchemical_validationtables will be pre-filled with empty values (and zeros in the validation columns) at onboarding for every site. One row should be created for each code. Thecc_mixtureandapplied_chemicalstable will not be prefilled. Rows will be added at parsing. The data flow team needs to be diligent to delete obsolete rows when a cover crop specie or chemical was mentioned by the field team by error, and then removed.
Relational Diagram:
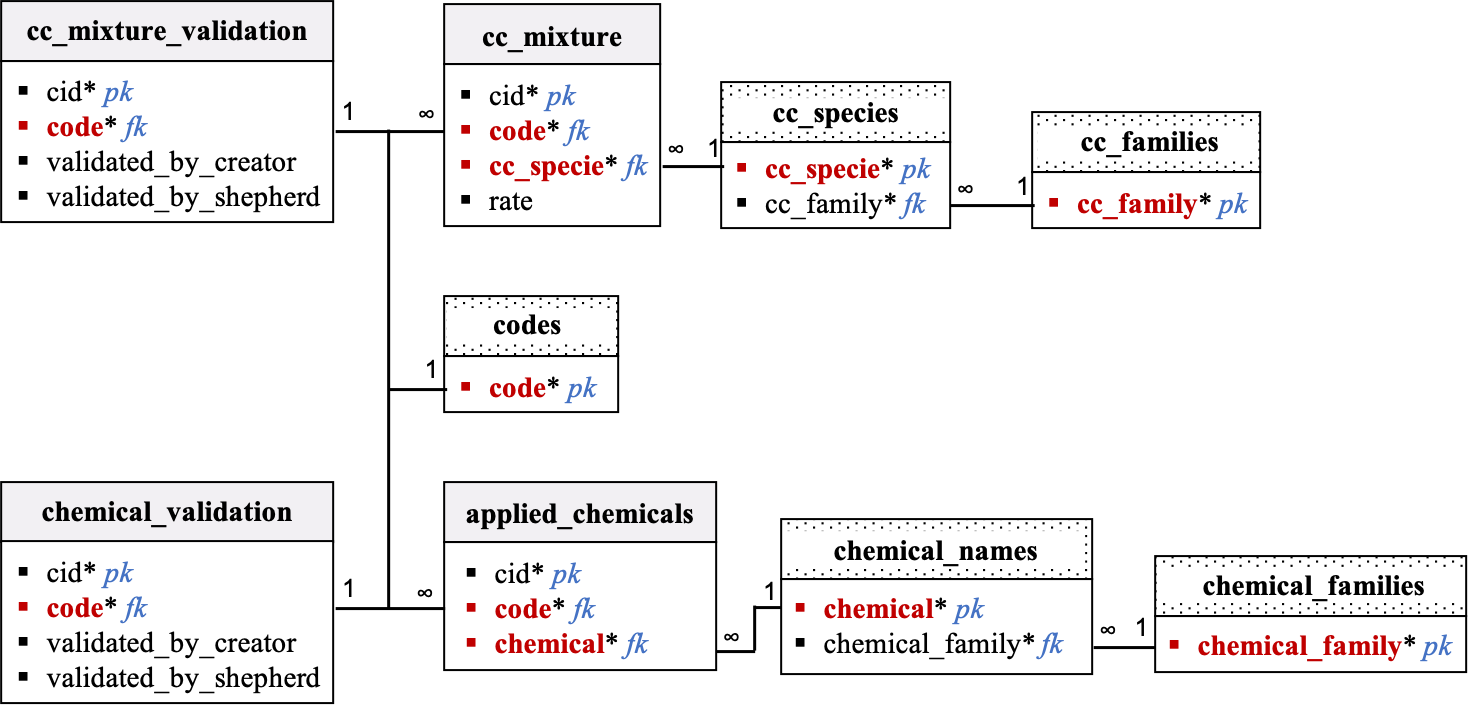
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
cc_families:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cc_family | charvar(20) | NA | Lists all cover crop families. |
cc_species:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cc_specie | charvar(20) | NA | Lists all cover crop species. |
| cc_family | charvar(20) | NA | Associates cover crop species to cover crop family. |
cc_mixture:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| cc_specie | charvar(20) | NA | Cover crop species planted in site. |
| rate | integer | lb/ac | Seeding rate of corresponding cover crop specie. |
cc_mixture_validation:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
chemical_families:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| chemical_family | charvar(30) | NA | Lists all chemical families. |
chemical_names:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| chemical | charvar(50) | NA | Lists all chemicals. |
| chemical_family | charvar(30) | NA | Associates chemicals to chemical family. |
applied_chemicals:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| chemical | charvar(50) | NA | Chemical applied to cover crop in site. |
chemical_validation:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
-
In-Field Biomass
Description:
-
The
biomass_in_fieldtable records the fresh biomass data collected in the field and data in these tables will be collected using Kobo. There are validation columns in this table. The field team will be able to suggest edits in the tech dashboard and the designated data shepherd will update data as needed. GIT issue tracker will be used in the background to keep track of suggestions and data updates. -
The
biomass_nirtable contains results of the NIR lab analysis conducted on the fresh biomass samples. Results from the NIR lab will be uploaded into the database via an upload functionality in the tech dashboard. There are validation columns in this table. Data should not need to be updated after import, provided that the uploaded file is in the proper format and there was no mistakes with sample ids. -
Rows in the
biomass_in_fieldandbiomass_nirtables will be pre-filled with empty values (and zeros in the validation columns) at onboarding when the in_field_biomass protocol is selected for a given site. Rows will be pre-filled with missing values (including values in the validation columns) if the in_field_biomass protocol is not selected at onboarding. 2 rows should be created in each table for each site code, whether or not the in_field_biomass protocol is selected at onboarding. -
Data validation will be performed for each observation/row in the data tables. After validation by the data creator and data shepherd, values in the validation_by_creator and validation_by_shepherd columns are set to 1. If the
biomass_in_fielddata are validated by both the data creator and the data shepherd while some values are still missing, then all missing values will be converted to -999. If allbiomass_in_fielddata is missing for a given site or if NIR analysis will not be conducted on a particular sample, then all values are converted to -999 and values in the validation_by_creator and validation_by_shepherd columns for that code are set to -999. -
legumes_40: Visual appreciation of the percentage of legumes in the cover crop. Equals 1 if legumes represent more than 40% of the cover crop, 0 otherwise. This information is necessary for the cover crop analysis.
Relational Diagram:
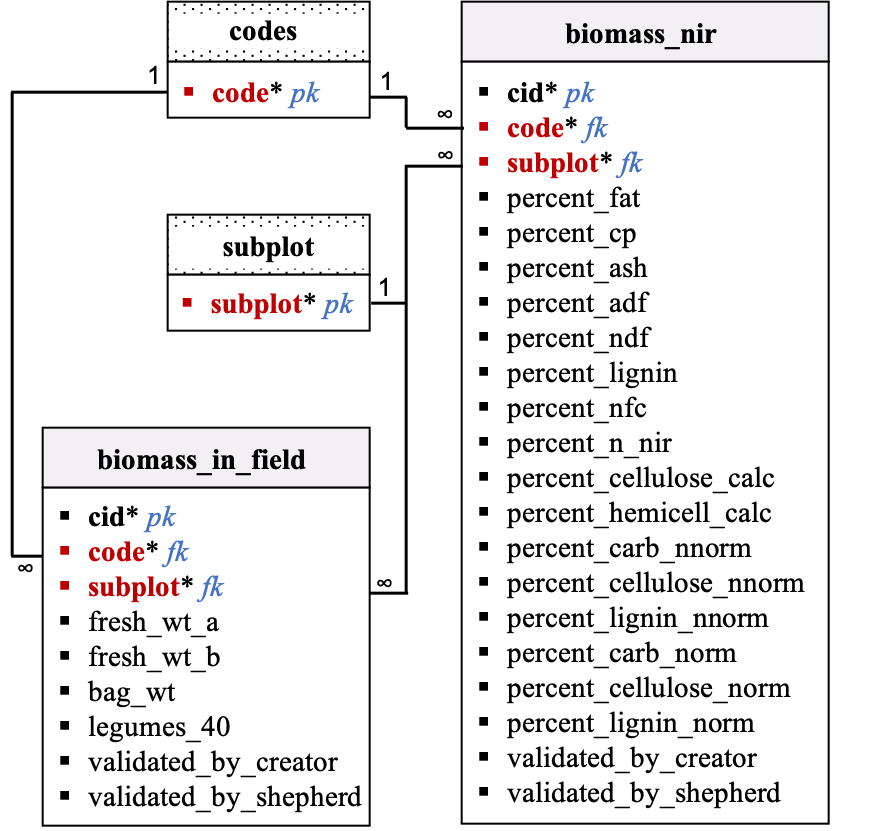
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
subplot:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subplot | integer | NA | List all subplots. |
biomass_in_field:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| fresh_wt_a | real | g | Fresh weight of sample A. |
| fresh_wt_b | real | g | Fresh weight of sample B. |
| bag_wt | real | g | Empty bag weight. |
| legumes_40 | integer | NA | Is there more than 40% of legumes in cover crop? |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
biomass_nir:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| percent_fat, percent_cp, percent_ash, percent_adf, percent_ndf, percent_lignin, percent_nfc | real | % | Results from NIR analysis. |
| percent_n_nir, percent_cellulose_calc, percent_hemicellulose_calc | real | % | Calculated results from NIR analysis . |
| percent_carb_nnorm, percent_cellulose_nnorm, percent_lignin_nnorm | real | % | Non-normalized results from NIR analysis. |
| percent_carb_norm, percent_cellulose_norm, percent_lignin_norm | real | % | Normalized results from NIR analysis. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
-
Decomp Biomass
Description:
-
The
decomp_biomass_freshanddecomp_biomass_drytables records cover crop biomass decomposition data collected after cover crop termination. Thedecomp_biomass_cnrecords cover crop C/N fraction data measured by our on-farm team in the lab. Thedecomp_biomass_ashtable records the cover crop ashless weights measured by our on-farm team in the lab. Data in all these tables will be collected using Kobo. There are validation columns in each of these four tables. The field team will be able to suggest edits in the tech dashboard and the designated data shepherd will update data as needed. GIT issue tracker will be used in the background to keep track of suggestions and data updates. -
Rows in the
decomp_biomass_fresh,decomp_biomass_dry,decomp_biomass_cn, anddecomp_biomass_ashtables will be pre-filled with empty values (and zeros in the validation columns) when the decomp_biomass protocol is selected for at onboarding. Rows will be pre-filled with missing values (including values in the validation columns) if the decomp_biomass protocol is not selected at onboarding. 20 rows should be created in each table for each site code. -
Data validation will be performed for each observation/row in the data table. After validation by the data creator and data shepherd, values in the validation_by_creator and validation_by_shepherd columns are set to 1. If data are validated by both the data creator and the data shepherd while some values are still missing, then all missing values will be converted to -999. If all data are missing for a given sample then all values for that sample and table are converted to -999 and values in the validation_by_creator and validation_by_shepherd columns for that code are set to -999.
Relational Diagram:
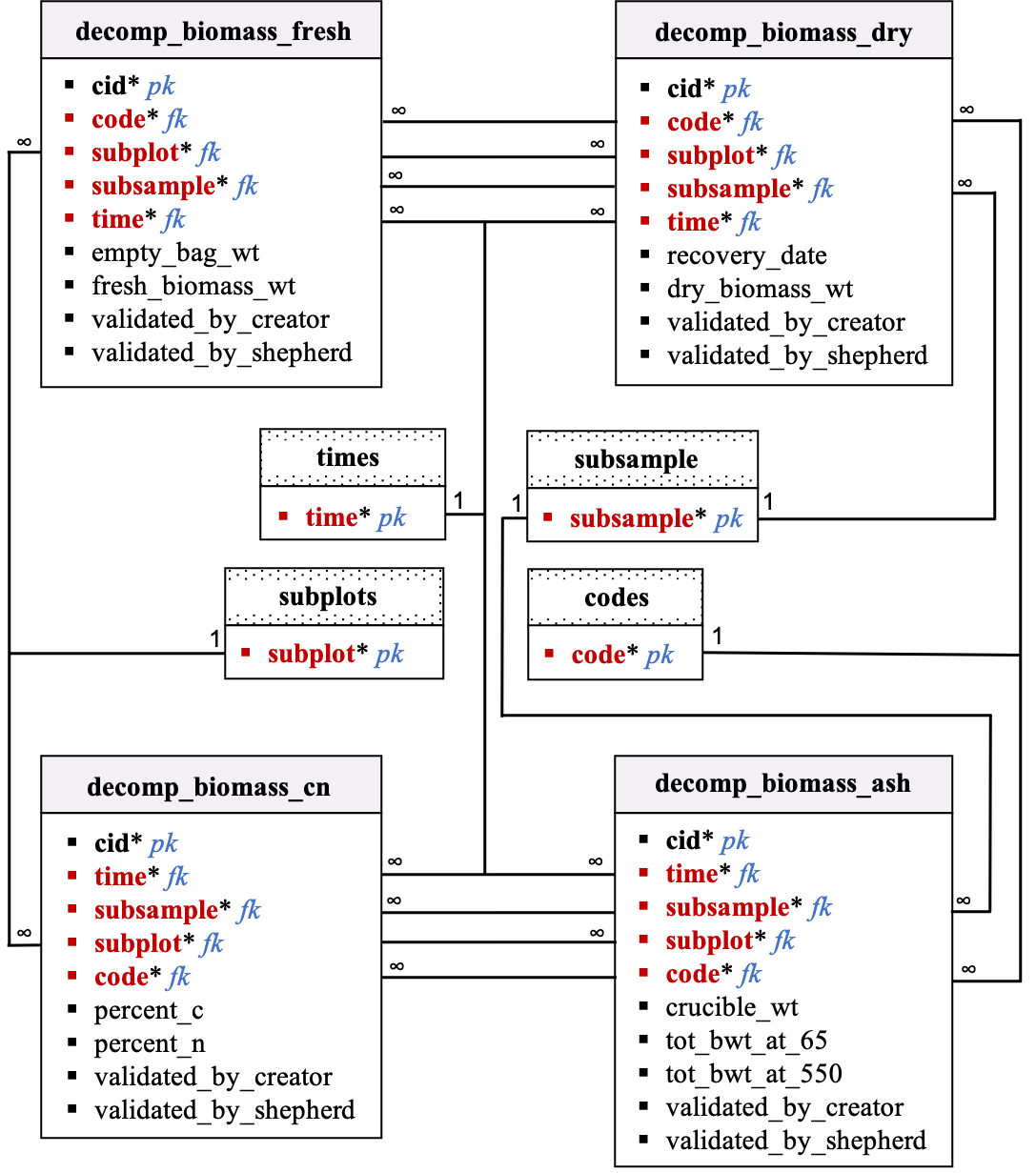
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
subplot:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subplot | integer | NA | List all subplots. |
subsamples:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subsample | char(1) | NA | List all subsamples. |
times:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| time | integer | NA | List all numeric/relative decomp bag pickup time. |
decomp_biomass_fresh:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| subsample | char(1) | NA | Subsample in subplot. |
| time | integer | NA | Numeric/relative decomp bag pickup time. |
| empty_bag_wt | real | g | Empty bag weight. |
| fresh_biomass_wt | real | g | Fresh biomass plus bag weight. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
decomp_biomass_dry:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| subsample | char(1) | NA | Subsample in subplot. |
| time | integer | NA | Numeric/relative decomp bag pickup time. |
| recovery_date | date | NA | Decomp bag recovery date. |
| dry_biomass_wt | real | g | Dry biomass plus bag weight. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
decomp_biomass_cn:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| subsample | char(1) | NA | Subsample in subplot. |
| time | integer | NA | Numeric/relative decomp bag pickup time. |
| percent_c | real | % | Percent carbon in decomposing cover crop residue. |
| percent_n | real | % | Percent nitrogen in decomposing cover crop residue. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
decomp_biomass_ash:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| subsample | char(1) | NA | Subsample in subplot. |
| time | integer | NA | Numeric/relative decomp bag pickup time. |
| crucible_wt | real | g | Weight of crucible fraction in cover crop residue. |
| tot_bwt_at_65 | real | g | Total biomass weight after drying at 65oC. |
| tot_bwt_at_550 | real | g | Total biomass weight after drying at 550oC. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
-
Soil Texture from Samples
Description:
-
The
texture_from_sampletable records soil texture measured at the TDR sensors from 0 to 30 cm, 30 to 60 cm, and 60 to 100 cm. Texture will be evaluated by an external laband results will be uploaded into the database via an upload functionality in the tech dashboard. There are two validation columns in this table. Data should not need to be updated after import, provided that the uploaded file is in the proper format. -
Rows in the
texture_from_samplestable will be pre-filled with empty values (and zeros in the validation columns) when the soil_texture protocol is selected at onboarding. Rows will be pre-filled with missing values (including values in the validation columns) when the soil_texture protocol is not selected at onboarding. 6 rows should be created in each table for each site code. -
Data validation will be performed for each observation/row in the data table. After validation by the data creator and data shepherd, values in the validation_by_creator and validation_by_shepherd columns are set to 1. If texture data is missing for a given site and subplot then all values are converted to -999 and values in the validation_by_creator and validation_by_shepherd columns for that code are set to -999.
-
Every words of the character strings inputed in the tclass column are be capitalized.
Relational Diagram:
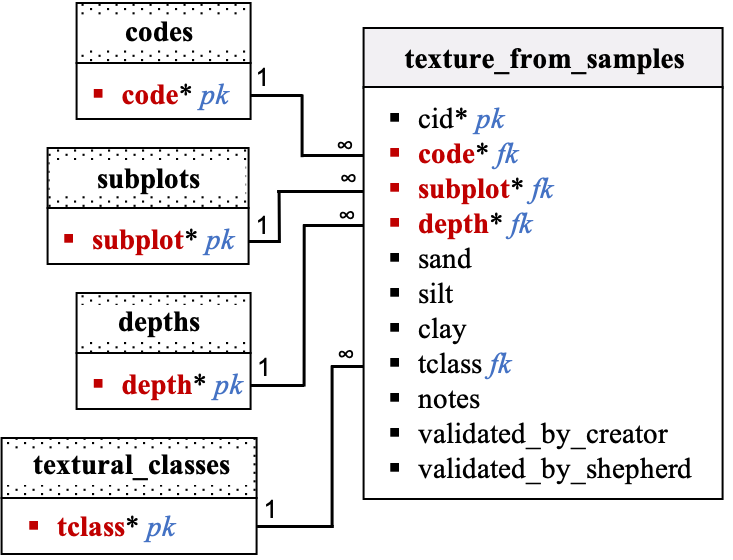
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
subplot:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subplot | integer | NA | List all subplots. |
depths:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| depth | integer | NA | List all soil sampling depths levels. |
textural_classes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| tclass | charvar(25) | NA | List all soil texture classes (USDA classification). |
texture_from_samples:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| depth | integer | NA | Depth level. |
| sand | real | % | Percent sand in soil. |
| silt | real | % | Percent silt in soil. |
| clay | real | % | Percent clay in soil. |
| tclass | charvar(25) | NA | Sample texture class. |
| notes | text | NA | Optional notes. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
-
Cash Crop Yield
Description:
-
The
yield_in_fieldtable records the row spacing and plant population data observed in the field. This data is necessary to calculate final plant population and cash crop yield. Population counts will only be made for corn since cotton and soybean have the ability to compensate for missing plants and lower populations (to a certain extent). Data in this table will be collected using Kobo. Theyield_corn,yield_soybeans, andyield_cottonrecords the yield data collected in the field and measured in the lab. Data in this tables will be collected using Kobo. There are validation columns in these four table. The field team will be able to suggest edits in the tech dashboard and the designated data shepherd will update data as needed. GIT issue tracker will be used in the background to keep track of suggestions and data updates. -
Rows in the
yield_in_fieldtable will be pre-filled with empty values (and zeros in the validation columns) when the cash_crop_yield protocol is selected at onboarding. Rows in theyield_corn,yield_soybeans, andyield_cottontables will be pre-filled accordingly to the specified cash crop. Rows will be pre-filled with missing values (including values in the validation columns) when the cash_crop_yield protocol is not selected at onboarding. -
The data flow teams needs to think about the steps to be taken if someone entered the wrong cash crop at onboarding. Then, the pre-filled row will have to be deleted in the yield table corresponding to the former cash crop, and a new row will have to be created in the yield table corresponding to the newly specified cash crop.
-
Data validation will be performed for each observation/row in the data tables. After validation by the data creator and data shepherd, values in the validation_by_creator and validation_by_shepherd columns are set to 1. If the cash crop yield data are validated by both the data creator and the data shepherd while some values are still missing, then all missing values will be converted to -999. If all data is missing for a given site and table then all values are converted to -999 and values in the validation_by_creator and validation_by_shepherd columns for that code are set to -999.
Relational Diagram:
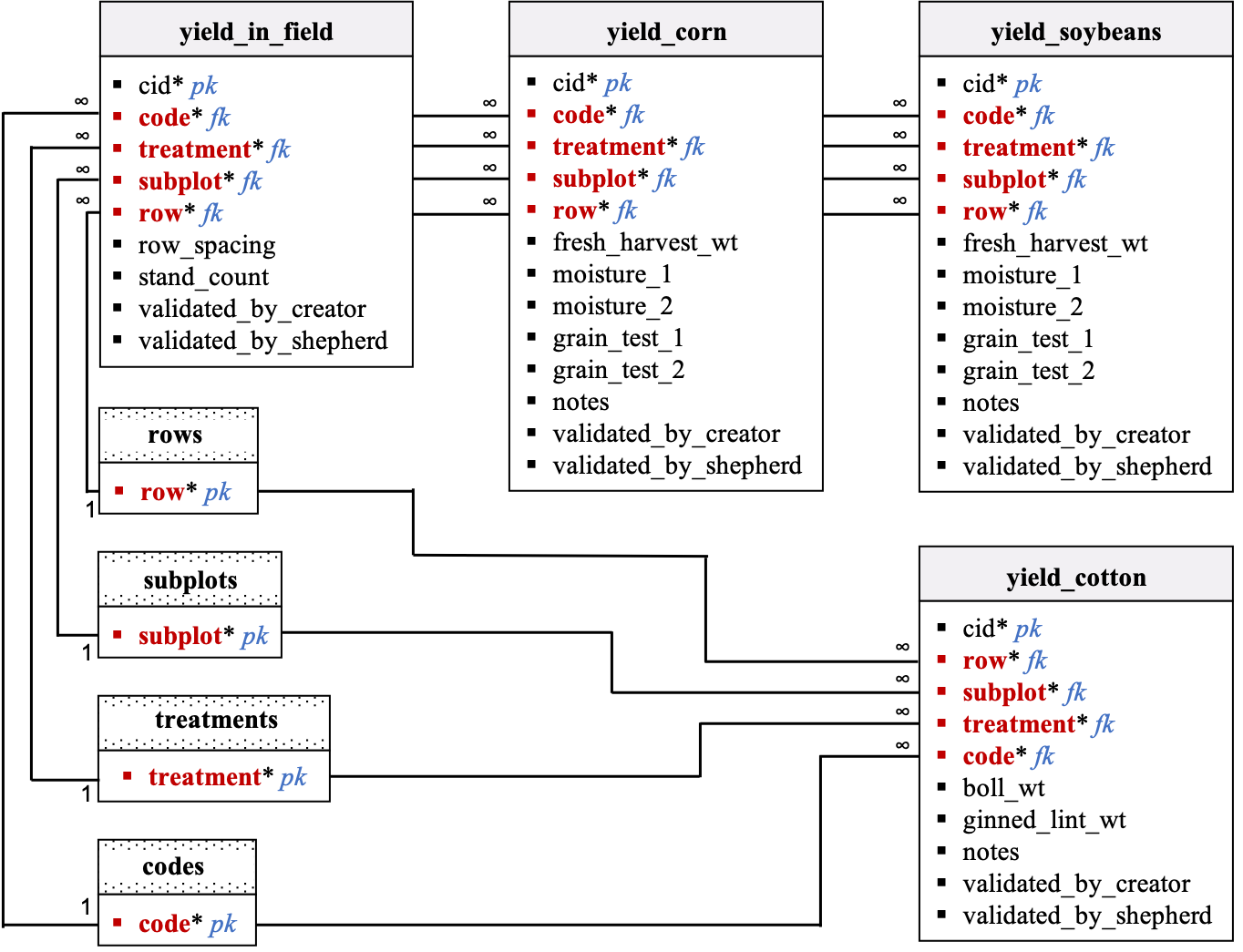
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
treatments:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| treatment | char(1) | NA | Lists all treatments. |
subplot:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subplot | integer | NA | List all subplots. |
rows:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| row | char(2) | NA | List all rows. |
yield_in_field:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| treatment | char(1) | NA | Treatment in site. |
| subplot | integer | NA | Subplot in treatment. |
| row | char(2) | NA | Row in subplot. |
| row_spacing | real | cm | Cash crop row spacing. |
| stand_count | real | NA | Number of plant per 10 ft row length [corn only]. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
yield_corn:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| treatment | char(1) | NA | Treatment in site. |
| subplot | integer | NA | Subplot in treatment. |
| row | char(2) | NA | Row in subplot. |
| fresh_harvest_wt | real | kg | Fresh grain/seed weight. |
| moisture_1 | real | % | Test moisture content, measurement 1. |
| moisture_2 | real | % | Test moisture content, measurement 2. |
| grain_test_1 | real | lb/bu | Grain test weight, measurement 1. |
| grain_test_2 | real | lbs/bu | Grain test weight, measurement 2. |
| notes | text | NA | Optional notes. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
yield_soybeans:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| treatment | char(1) | NA | Treatment in site. |
| subplot | integer | NA | Subplot in treatment. |
| row | char(2) | NA | Row in subplot. |
| fresh_harvest_wt | real | kg | Fresh grain/seed weight. |
| moisture_1 | real | % | Test moisture content, measurement 1. |
| moisture_2 | real | % | Test moisture content, measurement 2. |
| grain_test_1 | real | lb/bu | Grain test weight, measurement 1. |
| grain_test_2 | real | lbs/bu | Grain test weight, measurement 2. |
| notes | text | NA | Optional notes. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
yield_cotton:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| treatment | char(1) | NA | Treatment in site. |
| subplot | integer | NA | Subplot in treatment. |
| row | char(2) | NA | Row in subplot. |
| boll_wt | real | g | Cotton boll weight. |
| ginned_lint_wt | real | g | Cotton ginned lint weight. |
| notes | text | NA | Optional notes. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
-
GPS Locations
Description:
-
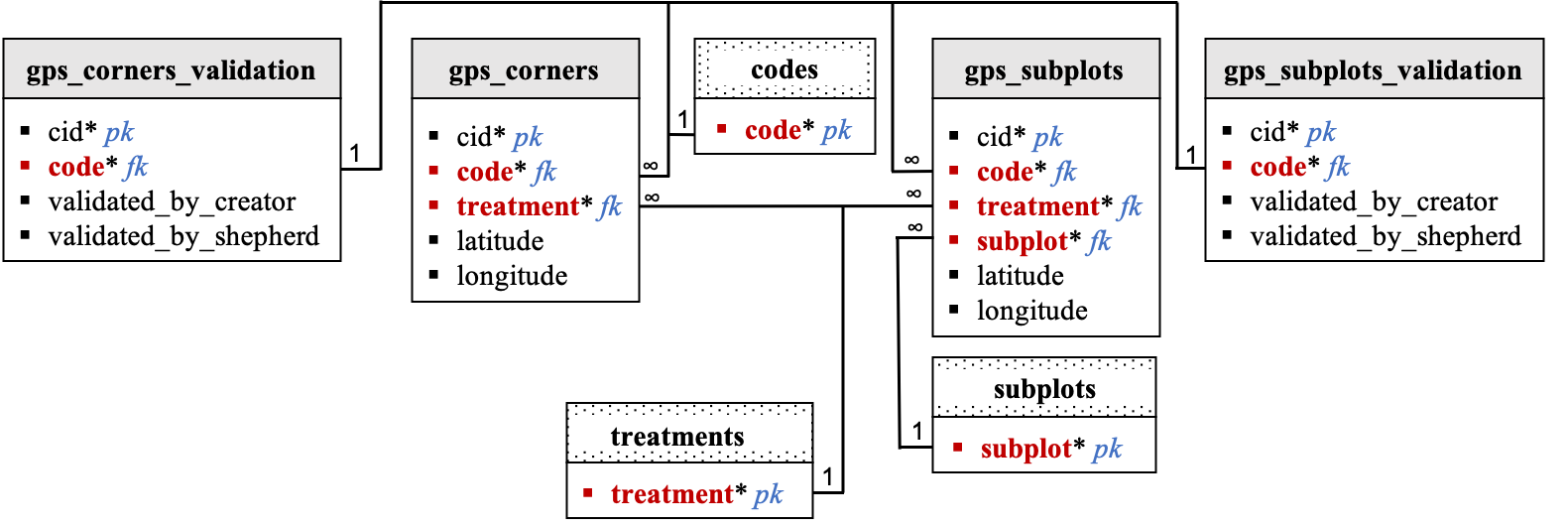
The
gps_cornersandgps_corners_validationtables records the GPS locations of the 4 cover crop and bare treatment corners in each site. Thegps_subplotsandgps_subplots_validationtables record the GPS locations of the TDR sensors placed in the cover crop and bare treatments in each site. The information will be gathered using Kobo. GPS data will be visibile on a map from the tech dashboard.After data collection, the field team will be able to suggest edits in the tech dashboard and the designated data shepherd will update data as needed. GIT issue tracker will be used in the background to keep track of suggestions and data updates. -
Validation columns are associated with the
gps_cornersandgps_subplotstables. Because the number of points collected might vary between sites, validation of thegps_cornersandgps_subplotstables data is completed in separate tables:gps_corners_validationandgps_subplots_validation, respectively. Validation will be performed by site code. Only thegps_corners_validationandgps_subplots_validationtables will be pre-filled at onboarding. By default, values in the validation_by_creator and validation_by_shepherd columns are set to 0 if the gps_locations protocol is selected for particular site. Values in these columns are set to -999 otherwise. If at least one row/observation is recorded in thegps_cornersandgps_subplotstables for a given site and that information was validated by the data creator and shepherd, then values of the validation_by_creator and validation_by_shepherd columns for that particular site are set to 1. If data is missing for a particular site, and the missing data is being validated by the data creator and shepherd then values of the validation_by_creator and validation_by_shepherd columns for that site are set to -999. -
The data flow team will need to think about how to manage GPS points that were not being collected properly (update if possible, and delete obsolete coordinates).
Relational Diagram:
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
treatments:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| treatment | char(1) | NA | Lists all treatments. |
subplot:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subplot | integer | NA | List all subplots. |
gps_corners:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| treatment | char(1) | NA | Treatment in site. |
| latitude | real | decimal degree | Latitude of GPS point [WGS84]. |
| longitude | real | decimal degree | Longitude of GPS point [WGS84]. |
gps_corners_validation:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
gps_corners:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| treatment | char(1) | NA | Treatment in site. |
| subplot | integer | NA | Subplot in treatment. |
| latitude | real | decimal degree | Latitude of GPS point [WGS84]. |
| longitude | real | decimal degree | Longitude of GPS point [WGS84]. |
gps_subplots_validation:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
-
Sensor Data
-
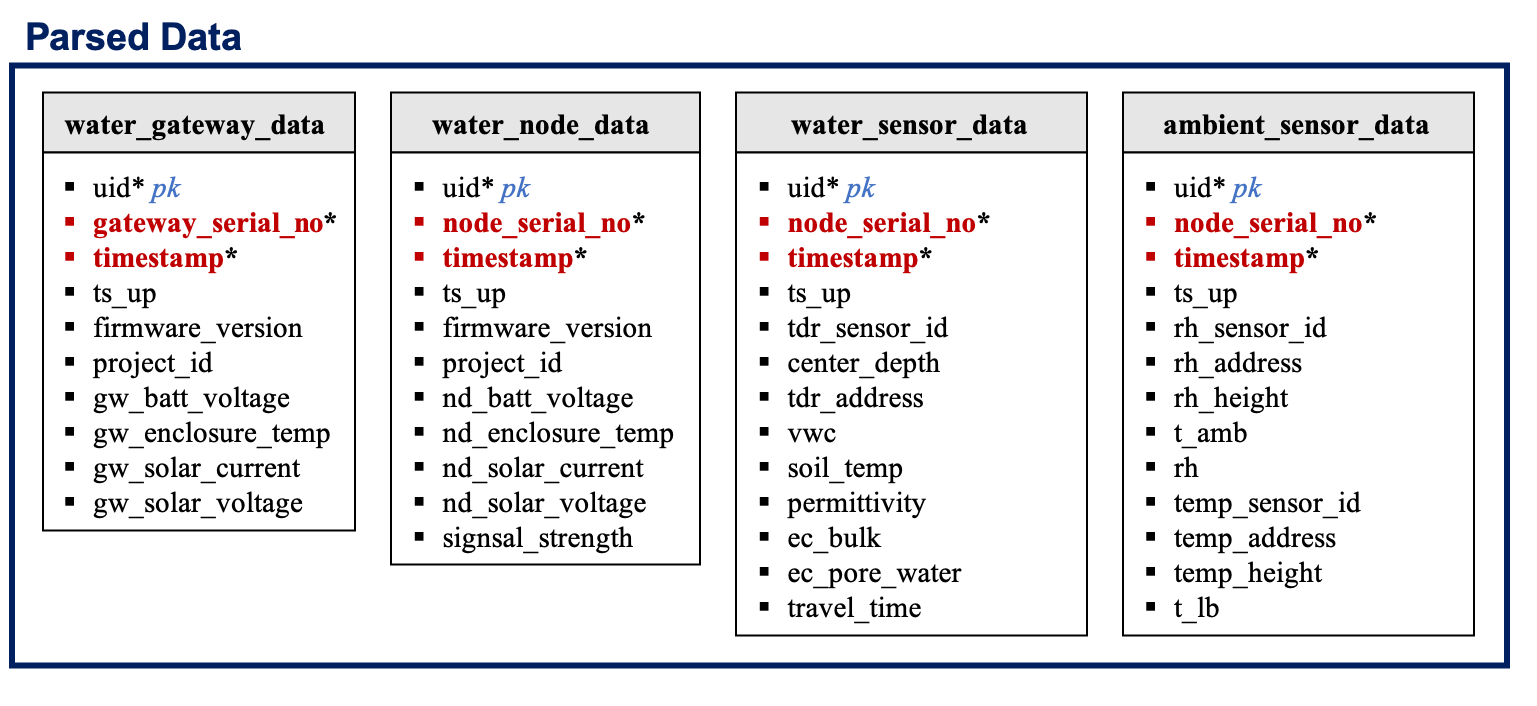
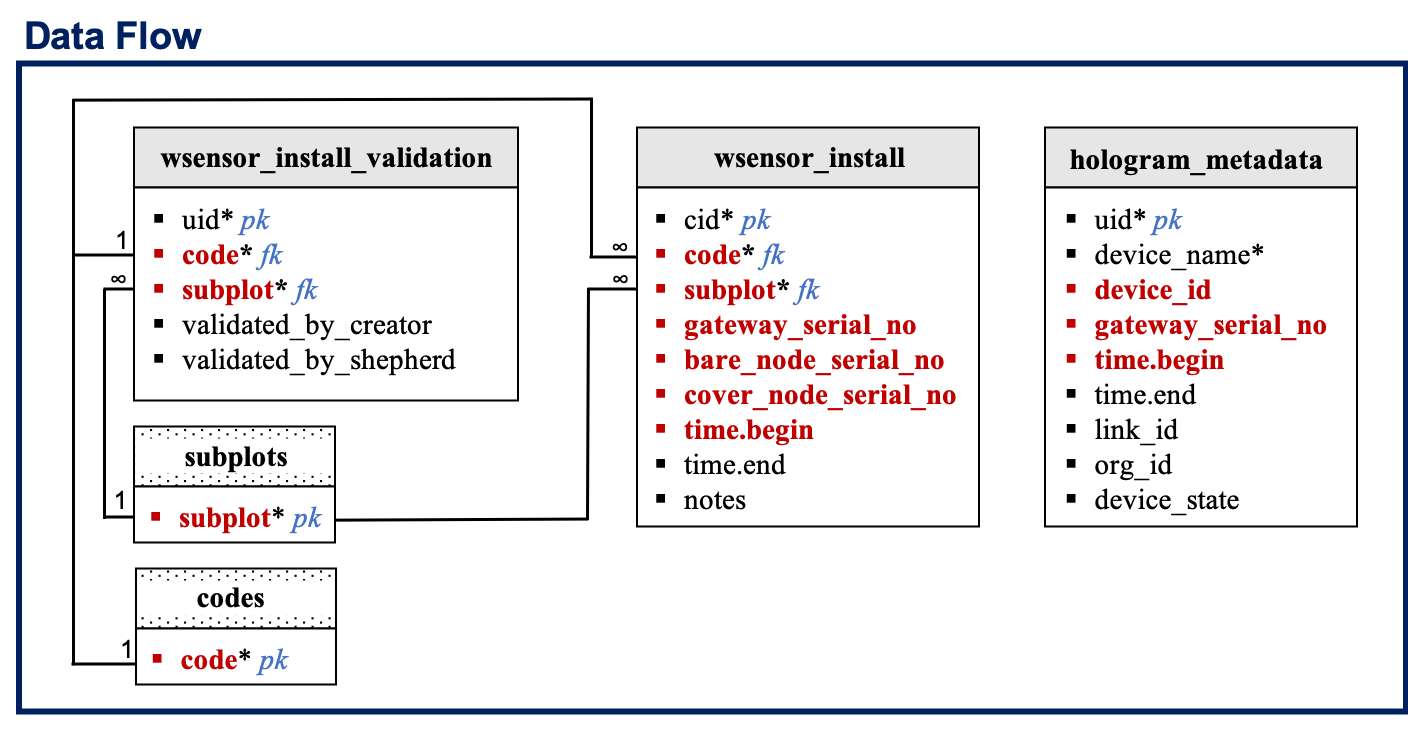
The
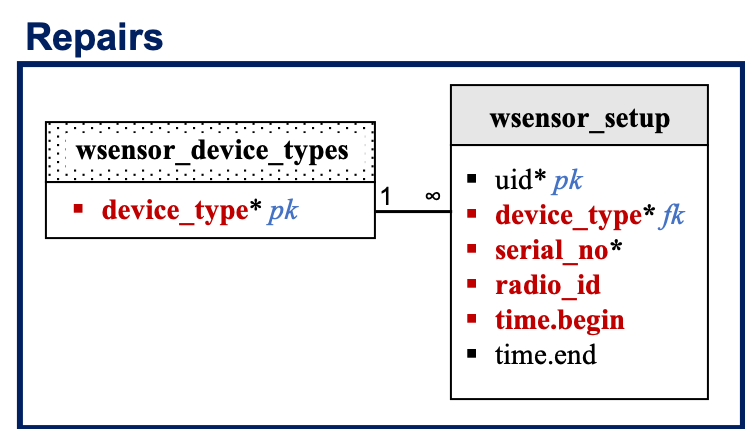
water_gateway_data,water_node_data,water_sensor_data, andambient_sensor_datatables record the hourly gateway, node, TDR sensor, and humidity plus temperature data, respectively. Data will be imported from hologram. If hologram fails, data will be downloaded and imported from the gateway SD card or node flash memory. Site codes, treatment, and subplot information are not being stored in these table. Allocation to a given site, treatment, and subplots will be made using thewsensor_installtable. Data in this table will be collected using Kobo. Only the sensor install data will be validated. Because material failure is possible, more than one observation/row can be entered for a given site code and validation will be conducted by site code in a separate table:wsensor_install_validation. Thehologram_devicestable lists the active devices in hologram and their unique hologram attributes. Thewsensor_setuptable will record the current device radio id to help address new node and gateways when equipment fails. -
The field team will be able to suggest edits only for the
wsensor_installandwsensor_setuptables. Edits will be suggested in the tech dashboard and the designated data shepherd will update data as needed. GIT issue tracker will be used in the background to keep track of suggestions and data updates. -
Only the
wsensor_install_validationtable will be pre-filled during onboarding. Validation will be performed by site code. By default, values in the validation_by_creator and validation_by_shepherd columns are set to 0 if the sensor_data protocol is selected for particular site, and -999 otherwise. If at least one row/observation is recorded in thewsensor_installtable for a given site and that information was validated by the data creator and shepherd, then values of the validation_by_creator and validation_by_shepherd columns for that particular site are set to 1. If data is missing for a particular site, and the missing data is being validated by the data creator and shepherd then values of the validation_by_creator and validation_by_shepherd columns for that site are set to -999. -
Values in ts_up columns are null if data was not successfully uploaded in hologram and manually collected from the SD card or flash memory. Project ID is
PSAfor PSA on-farm trials. Uppercase values in tdr_address indicate data collected in the bare treatment while lowercase values indicate data collected in the cover crop treatment. Sensor center_depth values are negative. Sensor height values are positive. -
Additional information about the sensor data inventory can be found here: https://docs.google.com/spreadsheets/d/1AqQ0j0mfNxKNU8u3s0NMxRO_-8DVzPzMzpEgAvC7Uuk/edit#gid=2136347016
Relational Diagram:
Data dictionary by table:
water_gateway_data:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| uid | serial | NA | Unique row id (ignore). |
| gateway_serial_no | char(8) | NA | Gateway serial number, 8-digit barcode. |
| timestamp | timestamp | NA | Time at which data were recorded [GMT]. |
| ts_up | timestamp | NA | Time at which data were uploaded into hologram [GMT]. |
| firmware_version | char(11) | NA | Firmware version. |
| project_id | charvar(5) | NA | Project ID. |
| gw_batt_voltage | real | V | Gateway battery voltage. |
| gw_enclosure_temp | real | Celcius | Gateway enclosure temperature. |
| gw_solar_voltage | real | V | Gateway photovoltaic voltage. |
| gw_solar_current | real | mA | Gateway photovoltaic current. |
water_node_data:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| uid | serial | NA | Unique row id (ignore). |
| node_serial_no | char(8) | NA | Node serial number, 8-digit barcode. |
| timestamp | timestamp | NA | Time at which data were recorded [GMT]. |
| ts_up | timestamp | NA | Time at which data were uploaded into hologram [GMT]. |
| firmware_version | char(11) | NA | Firmware version. |
| project_id | charvar(5) | NA | Project ID. |
| nd_batt_voltage | real | V | Node battery voltage. |
| nd_enclosure_temp | real | Celcius | Node enclosure temperature. |
| nd_solar_voltage | real | V | Node photovoltaic voltage. |
| nd_solar_current | real | mA | Node photovoltaic current. |
| signal_strength | real | dbm | Signal strength of gateway-node communication. |
water_sensor_data:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| uid | serial | NA | Unique row id (ignore). |
| node_serial_no | char(8) | NA | Node serial number, 8-digit barcode. |
| timestamp | timestamp | NA | Time at which data were recorded [GMT]. |
| ts_up | timestamp | NA | Time at which data were uploaded into hologram [GMT]. |
| tdr_sensor_id | charvar(33) | NA | TDR sensor id. |
| center_depth | integer | cm | Depth at the center of the sensor. |
| tdr_address | char(1) | NA | TDR sensor address. |
| vwc | real | vol/vol | Volumetric soil water content. |
| soil_temp | real | Celcius | Soil temperature. |
| permittivity | real | NA | Soil permittivity. |
| ec_bulk | real | 10^(-6)S/cm | Bulk soil electro-conductivity. |
| ec_pore_water | real | 10^(-6)S/cm | Pore water soil electro-conductivity. |
| travel_time | numeric | ps | Travel time. |
ambient_sensor_data:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| uid | serial | NA | Unique row id (ignore). |
| node_serial_no | char(8) | NA | Node serial number, 8-digit barcode. |
| timestamp | timestamp | NA | Time at which data were recorded [GMT]. |
| ts_up | timestamp | NA | Time at which data were uploaded into hologram [GMT]. |
| rh_sensor_id | charvar(33) | NA | Humidity sensor ID. |
| rh_address | char(1) | NA | Humidity sensor address. |
| rh_height | integer | cm | Height of the humidity center. |
| t_amb | real | Celcius | Air temperature at the humidity sensor. |
| rh | real | % | Relative humidity. |
| temp_sensor_id | charvar(33) | NA | Temperature sensor ID. |
| temp_address | char(1) | NA | Temperature sensor addres. |
| temp_height | integer | cm | Height of the temperature sensor. |
| t_lb | real | Celcius | Temperature below decomp bag. |
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
subplot:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subplot | integer | NA | List all subplots. |
wsensor_install:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| gateway_serial_no | char(8) | NA | Gateway serial number. |
| bare_node_serial_no | char(8) | NA | Bare node serial number. |
| cover_node_serial_no | char(8) | NA | Cover crop node serial number. |
| time.begin | timestamp | NA | Time when gateway and nodes are paired [GMT]. |
| time.end | timestamp | NA | Time when gateway and nodes are unpaired [GMT]. |
| notes | text | NA | Optional notes. |
wsensor_install_validation:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| uid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| subplot | integer | NA | Subplot in site. |
| validated_by_creator | integer | NA | Validation by data creator. |
| validated_by_shepherd | integer | NA | Validation by data shepherd. |
hologram_metadata:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| uid | serial | NA | Unique row id (ignore). |
| device_name | text | NA | Device name in hologram. |
| device_id | numeric(10) | NA | Device id in hologram. |
| gateway_serial_no | char(8) | NA | Gateway serial number, 8-digit barcode. |
| time.begin | timestamp | NA | Time when gateway and sim card were paired [GMT]. |
| time.end | timestamp | NA | Time when gateway and sim card were unpaired [GMT]. |
| link_id | numeric(10) | NA | Link id in hologram. |
| org_id | numeric(10) | NA | Org id in hologram. |
| device_state | bool | NA | Device status in hologram. |
wsensor_device_types:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| device_type | charvar(15) | NA | Lists all possible device types. |
wsensor_setup:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| uid | serial | NA | Unique row id (ignore). |
| device_type | charvar(15) | NA | Device types. |
| serial_no | char(8) | NA | Device serial number, 8-digit barcode. |
| radio_id | charvar(2) | NA | Device radio ID. |
| time.begin | timestamp | NA | Time when gateway and radio ID were paired [GMT]. |
| time.end | timestamp | NA | Time when gateway and radio ID were unpaired [GMT]. |
-
Weather
Description:
-
The
rainfallandweathertable record hourly rainfall and atmospheric weather data. Weather data will be imported from our weather API. Data are stored in two separate tables because atmospheric weather data are not being updated by the data source as often as the rainfall data. There are no validation columns associated with the weather data. Data sources are being recorded within the data table to keep track of possible future changes in data source. -
Hourly weather data for any given site were recorded from August 1st of the previous year to December 31st of the cash crop year.
-
Wind speed can be calculated from the zonal_wind and meridional_wind columns using the following formula:
wind_speed [m/s] = sqrt(zonal_wind_speed^2 + meridional_wind_speed^2) -
Relative humidity can be calcuated from the humidity column using the following formula:
relative_humidity (((humidity/18)/(((1-humidity)/28.97) + (humidity/18)))*pressure/1000) /(0.61*exp((17.27*air_temperature)/(air_temperature + 237.3)))
Relational Diagram:

Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
rainfall:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| date | date | NA | Date (YYYY-MM-DD). |
| timez | timestamp | NA | Timestamp [GMT]. |
| rainfall | real | mm | Rainfall amount. |
| data_source_rainfall | charvar(10) | NA | Rainfall data source. |
weather:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| date | date | NA | Date (YYYY-MM-DD). |
| timez | timestamp | NA | Timestamp [GMT]. |
| air_temperature | real | Celcius | Air temperature. 2m above ground. |
| humidity | real | kg/kg | Specific humidity. 2m above ground. |
| longwave_radiation | real | W/m2 | Surface downward longwave radiation. |
| shortwave_radiation | real | W/m2 | Surface downward shortwave radiation. |
| zonal_wind | real | m/s | Zonal wind. 10m above ground. |
| meridional_wind | real | m/s | Meridional wind. 10m above ground. |
| pressure | real | Pa | Surface pressure. |
| convective_precipitation | real | NA | Fraction of total precipitation that is convective. |
| potential_energy | real | J/kg | Convective available potential energy. |
| potential_evaporation | real | kg/m2 | Potential evaporation. |
| data_source_atmosphere | charvar(10) | NA | Atmospheric weather data source. |
-
Weed Research
Description:
- Weed pictures will not be stored in this database and the
weed_researchtable was designed as a place holder which can used to facilitate linkage between weed pictures and other data collected at PSA on-farm sites. This table is presently empty. There are no validation columns in this table. Data in theweed_researchtable could be imported automatically with a script that automatically parse through the directories containing weed research pictures.
Relational Diagram:
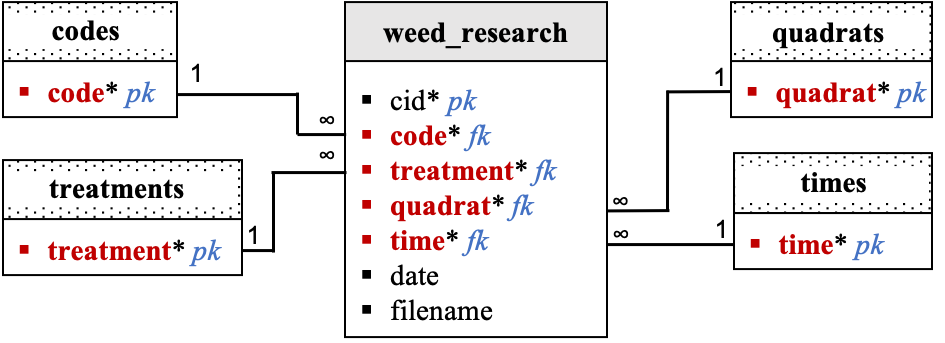
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
treatments:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| treatment | char(3) | NA | Lists all treatments. |
quadrats:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| quadrat | char(3) | NA | Lists all quadrats. |
times:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| time | integer | NA | List all numeric/relative data collection time. |
weed_research:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| treatment | char(1) | NA | Treatment in site. |
| quadrat | integer | NA | Quadrat in treatment. |
| time | integer | NA | Numeric/relative data collection time. |
| date | date | NA | Picture data collection date. |
| filename | charvar(30) | NA | Picture filename. |
-
Surface Bulk Density [Retired Protocol]
Description:
- The
bulk_densitytable contains surface bulk density data for the 2018 NC and MD sites. Data in this table are not being pre-filled at planting since the bulk_density protocol has been retired. There are no validation columns associated with this table.
Relational Diagram:
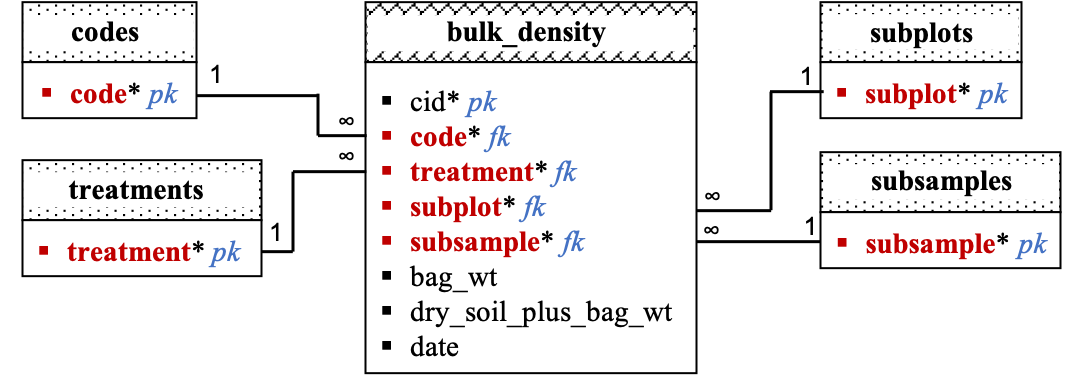
Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
treatments:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| treatment | char(3) | NA | Lists all treatments. |
subplots:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subplot | integer | NA | Lists all subplots. |
subsamples:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subsample | char(1) | NA | Lists all subsamples. |
bulk_density:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| treatment | char(1) | NA | Treatment in site. |
| subplot | integer | NA | Subplot in treatment. |
| subsample | char(1) | NA | Subsample in subplot. |
| bag_wt | real | g | Empty bag weight. |
| dry_soil_plus_bag_wt | g | Weight of dry soil sample plus bag. | |
| date | date | NA | Date of bulk density measurement. |
-
Soil Nitrogen [Retired Protocol]
Description:
- The
soil_nitrogentable contains soil nitrogen data for the 2017 and 2018 NC and MD sites. Data in this table are not being pre-filled at planting since the soil_nitrogen protocol has been retired. There are no validation columns associated with this table.
Relational Diagram:

Data dictionary by table:
codes:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(3) | NA | Lists all possible site codes. |
seasons:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| code | char(1) | NA | Lists all possible seasons. |
treatments:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| treatment | char(3) | NA | Lists all treatments. |
subplots:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| subplot | integer | NA | Lists all subplots. |
depths:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| depth | integer | NA | Lists all depths levels. |
soil_nitrogen:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| season | char(1) | NA | Season. |
| treatment | char(1) | NA | Treatment in site. |
| subplot | integer | NA | Subplot in treatment. |
| depth | integer | NA | Depth level in subplot. |
| empty_bag_wt | real | g | Empty bag weight. |
| soil_plus_bag_wt | real | g | Fresh soil plus bag weight. |
| rock_wt | real | g | Weight of rocks in sample. |
| tin_wt | real | g | Empty tin weight. |
| moist_soil_plus_tin_wt | real | g | Fresh soil in tin plus tin weight. |
| dry_soil_plus_tin_wt real | g | Dry soil in tin plus tin weight. | |
| n_vial_nb | charvar(3) | NA | Vial number for nitrogen analysis. |
| n_soil_dry_wt | real | g | Dry soil weight in vial. |
| nh4_ppm | charvar(10) | ppm | Concentration of NH4 in sample. |
| no3_ppm | charvar(10) | ppm | Concentration of NO3 in sample. |
farm_history [retired]:
| Columns | Data Type | Unit | Description |
|---|---|---|---|
| cid | serial | NA | Unique row id (ignore). |
| code | char(3) | NA | Site code. |
| fall_sampling_date | date | NA | Fall deep core sampling date. |
| spring_sampling_date | NA | Spring deep core sampling date. | |
| post_harvest_fertility | integer | NA | 1 if post harvest fertility was applied, 0 otherwise. |
| post_harvest_fertility_date | date | NA | Post-harvest fertility date. |
| post_harvest_fertility_source | text | NA | Post-harvest fertility source. |
| post_harvest_fertility_rate | text | NA | Post-harvest fertility rate. |
| at_pre_planting_fertilization | integer | NA | 1 if at/pre-planting fertilization was applied. |
| at_pre_planting_fertilization_date | date | NA | At/pre-planting fertilization date. |
| at_pre_planting_fertilization_method | text | NA | At/pre-planting fertilization method. |
| at_pre_planting_fertilization_rate | text | NA | At/pre-planting fertilization rate. |
| sidedress | integer | NA | 1 if cash crop was applied, 0 otherwise. |
| sidedress_date | date | NA | Sidedress date. |
| sidedress_rate | text | NA | Sidedress rate. |